BIRSBIO 2020 Hackathon
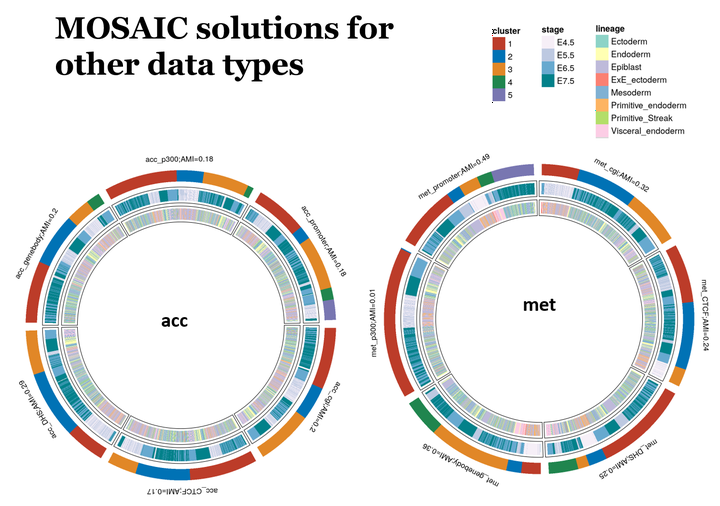
scNMT seq data encompasses rich information across the transcriptome and epigenome avenues capturing the stages of mouse gastrulation. This data set was challenging in terms of missingness and integration across various data types like RNA transcripts, DNA accessibility and DNA methylation, to understand their role independently and as a cross play with each other defining biological functions.
We wish to address the problem of identifying localized molecular signatures with respect to an outcome of interest, like stage and lineage. This poses an interesting challenge as in to understand heterogeneity in a population by not unsupervised clustering followed by hypothesis testing, but in a more targeted fashion where the cluster solutions are more meaningful and driven by outcome of interest
MOSAIC, or Multi Omic Supervised Integrative Clustering, is a supervised clustering algorithm, inspired from survClust. The primary advantage of such a supervised or weighted approach is that the final clusters are robust and more meaningful in terms of outcome of interest. Our approach downplays the unrelated features and only keeps the contribution from associated features, making it resistant to noise from other data trends. It also provides a novel approach to deal with samples that have missing data across data types, where in most cases the analysis would be reduced to samples with complete data. MOSAIC on the other hand can handle missing data and tries to retain maximum number of samples.
MOSAIC analysis results can be found here - https://github.com/arorarshi/scNMT_seq_MOSAIC